Testing the Water: DNA Sampling to Detect Water Contaminants
By Dan DeBaunShare
Water contamination by microbial contaminants is a widespread problem throughout the country. These contaminants make their way into freshwater systems, where they pose a health to humans. While water regulators routinely test water sources for these types of contaminants, the methods used are outdated and unreliable.
Now, researchers from the Lawrence Berkeley National Laboratory (Berkeley Lab) have found a way to not only accurately detect microbial contaminants in our waterways, but also to distinguish between different sources of those contaminants.
Using an award-winning device known as a PhyloChip, which is about the size of a credit card, and which can reliably detect more than 60,000 microbe species, water regulators can now test the waters with greater accuracy. During preliminary testing of the device on Northern California's Russian River watershed, the scientists found cases where this new identified microbes that posed a potential health risk to humans that had not been detected by conventional fecal count tests. The study, which was recently published in the scientific journal Water Science, also found that in some cases the conventional testing methods flagged bacteria that were not considered a health risk to humans.
"With the PhyloChip, in an overnight test we can get a full picture of the microorganisms in any given sample," said Eric Dubinsky, a microbial ecologist at Berkeley Lab and lead author of the paper. "Instead of targeting one organism, we're essentially getting a fingerprint of the microbial community of potential sources in that sample. So it gives us a more comprehensive picture of what's going on. It's a novel way of going about source tracking."
Local water regulators currently collect water samples, and then culture the bacteria for 12 hours before checking the levels of two key bacteria types: Enterococcus and E. Coli — which are considered indicators of fecal contamination. But this method fails to distinguish between different sources of the bacteria, which could have originated from cattle, waterfowl, humans, sewage or even rotting vegetation.
"These tests have been used for decades and are relatively primitive," Dubinsky said. "Back in the 1970s when the Clean Water Act was developed and we had sewage basically flowing into our waters, these tests worked really well. Epidemiological studies showed an association of these bacteria with levels of illness of people who used the water. These bacteria don't necessarily get you sick, but they're found in sewage and fecal matter. That's why they're measured."
While it is easy to identify point sources, such as sewage outfalls, which generally get cleaned up once they have been identified, non-point sources of pollution, such as runoff from agricultural lands, are more difficult to identify, and are becoming a growing concern.
The PhyloChip, which was developed by co-author, Gary Andersen, a microbial ecologist at Berkeley Lab, together several colleagues at Berkeley Lab, has proven useful for a number agricultural, medical, and environmental applications, including gaining a better understanding of coral reef ecology, air pollution, and environmental conditions in the Gulf of Mexico following the BP oil disaster. The PhyloChip has 1 million probes which enable it to identify microbes according to variations in a specific gene, without the need to culture the bacteria overnight. The scientists soon realized that the PhyloChip held great potential for assessing water quality and pinpointing the source of water contaminants.
It is no simple task to determine the source of a pathogen. In many cases, more than one type of microbe is needed to determine the source, as the microbial community of animals such as cows can consist of a thousand different microbial organisms.
To address this, the scientists coerced a lab intern into going around and collecting poop from a wide range of animals. They then set about cataloguing the microbial communities found in the poop specimens of cattle, horses, pigs, raccoons, sea lions, different bird species, as well as other wildlife, humans and sewage, using that catalogue to develop a model which compares an unknown microbial sample with the samples in their reference library.
"We've used the PhyloChip in a way that it hasn't been used before by using machine learning models to analyze the data in order to detect and classify sources," Andersen said. "It's essentially giving you a statistical probability that a microbial community came from a particular source."
After comparing their method with forty others used to track sources of microbial contaminants, their method proved to be the only one that could reliably detect all microbial sources correctly. Even when the microbes originate from an animals source that is not listed in their library catalogue, this method can still prove to be extremely useful in identifying the source. For example, in one study the sample was from a chicken, but the team had not yet analyzed chickens. However, they did have records of pigeons, gulls and geese, which enabled them to determine that the sample came from a bird.
After extensive sampling within the Russian River watershed, which currently does not comply with the Clean Water Act, the scientists discovered that contamination from human sources was widespread around areas where communities depend largely on aging septic tank systems. They also detected significant contamination from human sources following a weekend jazz concert, which was not as clearly evident when using the other methods. Dubinsky attributes this to the fact that this new methods is much more sensitive to human contaminants that the outdated fecal indicator tests.
The Berkeley Lab scientists are currently working together with the EPA to develop the method further so that it can be used universally at any location, by anyone, even non-experts. The method also holds promise for determining the sources of nutrients that fuel algal blooms, particularly in the Great Lakes, where this continues to be an ongoing problem.
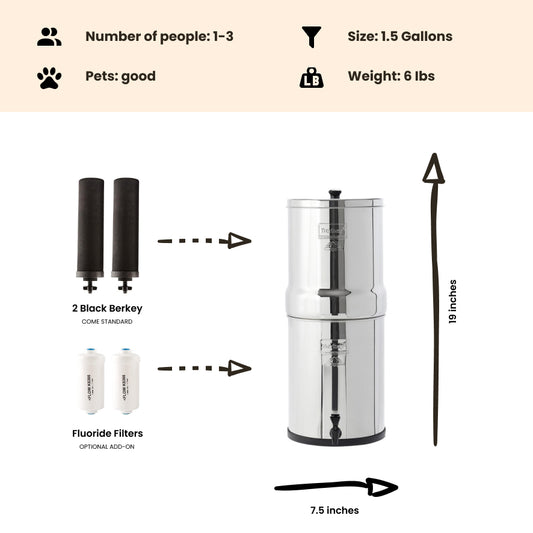
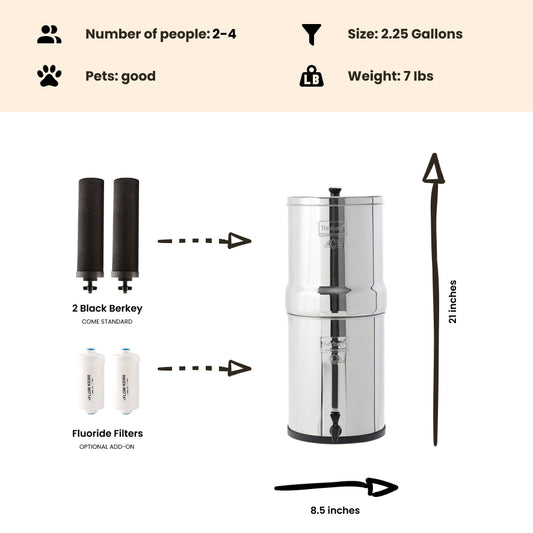
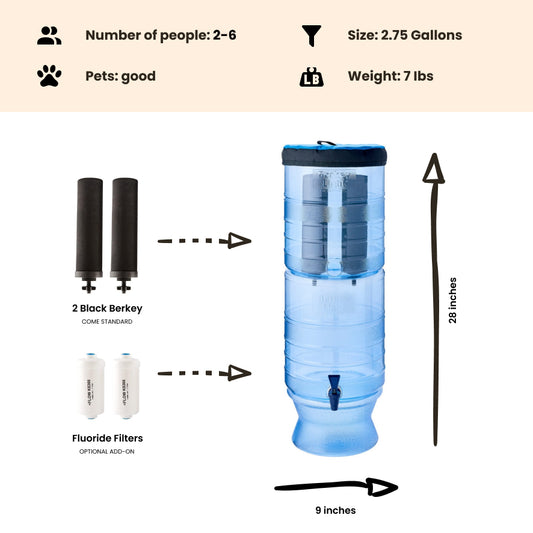
If you're concerned your water may be contaminated, the Berkey systems equipped with the black berkey filters will remove bacteria and viruses to levels greater than 99.9999%
Journal Reference:
Eric A. Dubinsky, Steven R. Butkus, Gary L. Andersen. Microbial source tracking in impaired watersheds using PhyloChip and machine-learning classification. Water Research, 2016; 105: 56 DOI: 10.1016/j.watres.2016.08.035
-
Regular price From $302.00 USDRegular priceUnit price / per
-
Regular price $234.00 USDRegular priceUnit price / per
-

 Sold outRegular price From $305.00 USDRegular priceUnit price / per
Sold outRegular price From $305.00 USDRegular priceUnit price / per -
Regular price $327.00 USDRegular priceUnit price / per
-
Regular price From $367.00 USDRegular priceUnit price / per
-
Regular price From $408.00 USDRegular priceUnit price / per
-
Regular price From $451.00 USDRegular priceUnit price / per

Dan DeBaun
Dan DeBaun is the owner and operator of Big Berkey Water Filters. Prior to Berkey, Dan was an asset manager for a major telecommunications company. He graduated from Rutgers with an undergraduate degree in industrial engineering, followed by an MBA in finance from Rutgers as well. Dan enjoys biohacking, exercising, meditation, beach life, and spending time with family and friends.
~ The Owner of Big Berkey Water Filters